
This web site, https://www.vcru.wisc.edu/sdata, contains data from the laboratory of Philipp W. Simon, USDA-ARS Vegetable Crops Research Unit [Click here for our web page]
 |
USDA ARS VCRU Data Server This web site, https://www.vcru.wisc.edu/sdata, contains data from the laboratory of Philipp W. Simon, USDA-ARS Vegetable Crops Research Unit [Click here for our web page] |
The following table is from Simon, P.W., Plant Breeding Reviews (2000) 19:157-190. (Slightly modified)
| Gene symbolz |
Character description |
Gene source | Reference |
|---|---|---|---|
| A | a-Carotene synthesis (may be identical to lo or O) |
'Kintoki' | Umiel and Gabelman 1972 |
| (Ce) | Cercospora leaf spot resistance | WCR-1 Wisconsin inbred | Angell and Gabelman 1968 |
| (Cr) | Cracking roots (dominant to non-cracking) | 'Touchon' | Dickson 1966 |
| Eh | Downy mildew (Erysiphe heraclei) resistance | D. carota ssp. dentatus | Bonnet 1983 |
| g | g = Green petiole, G = Purple petiole | 'Tendersweet' | Angell and Gabelman 1970 |
| gls | Glabrous seedstalk | W-93 Wisconsin inbred | Morelock and Hosfield 1976 |
| lo | Intense orange xylem | Miscellaneous | T.Kust, 1970 (cited in Buishand and Gabelman, 1979) |
| L | Lycopene synthesis | 'Kintoki' | Umiel and Gabelman 1972 |
| (mh-1), (mh-2) | Meloidogyne hapla resistance | 'Rotin', Wisconsin inbreds | Wang and Goldman 1996 |
| Mj-1 | Meloidogyne javanica, resistance | 'Brasilia' | Simon et al. 2000 |
| Ms-1, Ms-2, Ms-3 | Maintenance of male sterility | 'Tendersweet' | Thompson 1962 |
| Ms-4, ms-5 | 'Tendersweet', 'Imperator 58', PI 169486 | Hansche and Gabelman 1963 | |
| O | Orange xylem | Miscellaneous | T.Kust, 1970 (cited in Buishand and Gabelman, 1979) |
| P-1 | Purple root | PI 173687 | Simon 1996 |
| P-2 | Purple node | PI 175719 | Simon 1996 |
| (P-3), (P-4) | Purple root | Miscellaneous | Laferriere and Gabelman 1968 |
| rp | Reduced carotenoid pigmentation | W266 Wisconsin inbred | Goldman and Breitbach 1996 |
| rs | Reducing sugar in root | Miscellaneous | Freeman and Simon 1983 |
| (sp-1), (sp-2) | Seed spine formation | 'Amkaza' | Nieuwhof and Garritsen 1984 |
| y | Yellow xylem | Miscellaneous | T.Kust, 1970 (cited in Buishand and Gabelman 1979) |
| Y-1 | Differential xylem / phloem carotene levels | Miscellaneous | T.Kust, 1970 (cited in Buishand and Gabelman 1979) |
| Y-2 | Differential xylem / phloem carotene levels | Miscellaneous | T.Kust, 1970 (cited in Buishand and Gabelman 1979) |
z Loci enclosed in parentheses were not named previously; suggested symbol. Isozyme and DNA markers not included.
| Gene Symbol | Phenotype | Source | Reference |
|---|---|---|---|
| A | a-carotene synthesis | Kintoki | Umiel and Gableman, 1986 |
| Io | Intense orange xylem | Multiple | Kust, 1971 |
| L | Lycopene synthesis | Kintoki | Umiel and Gableman, 1986 |
| O | Orange xylem | Multiple | Kust, 1971 |
| P1 | Purple root | Multiple | Simon, 1996 |
| y | Yellow xylem | Multiple | Laferierre and Gabelman, 1968 |
| Y1 | Differential xylem and phloem pigmentation | Multiple | Kust, 1971 |
| Y2 | Yellow xylem and phloem1 | Multiple yellow and white carrots | Kust, 1971 |
| Rp | Reduced pigment | W266D | Goldman and Breitbach, 1996 |
1in some backgrounds phloem will be orange. Table adapted from Peterson and Simon (1986).
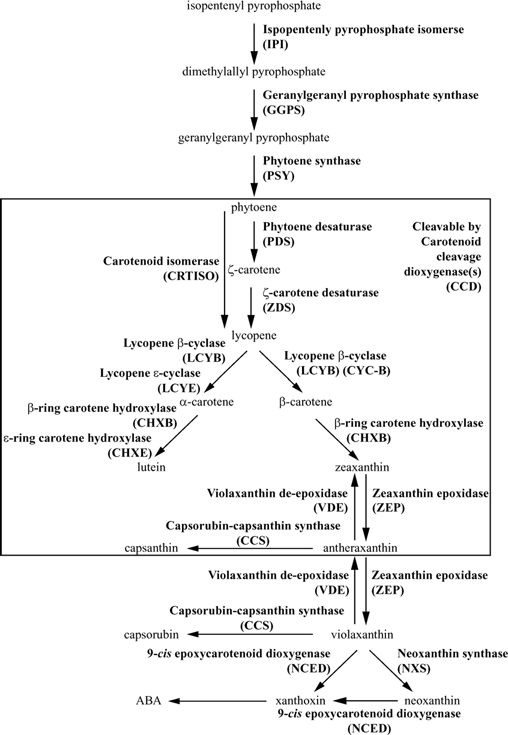
 Pathways
Pathways
The following figure is from Brian Just, Ph.D. Thesis, University of Wisconsin-Madison, 2004, page 25, Figure 1.1
A simplified diagram showing the enzymes and major products of the carotenoid pathway, parts of which are common to all plants. The product names appear between the arrows and are written in regular type. The enzyme names are written in boldface with the abbreviation used throughout this thesis below in parentheses. Products from phytoene through violaxanthin plus neoxanthin are carotenoids. Phytoene, z-carotene, lycopene, b-carotene, and a-carotene are carotenes. Lutein, zeaxanthin, antheraxanthin, violaxanthin, capsanthin, capsorubin, and neoxanthin are xanthophylls. Xanthoxin and the plant hormone ABA are apocarotenoids. The products appearing in the large box can be cleaved into apocarotenoid products by a class of enzymes called carotenoid cleavage dioxygenases (CCDs). Figure compiled and summarized from Liu et al., 2004; Giuliano et al., 2000; Hirschberg, 2001; Cunningham and Gantt, 1998.
This is a clickable pathway, click on the enzyme name to view more information
This enzymes catalyzes the conversion of geranylgeranyl pyrophosphate to phytoene
One copy of the IPI gene has been found:
IPI could not be mapped because no polymorphisms were found
This enzymes catalyzes the conversion of phytoene to z-carotene (zeta-carotene)
Two copies of the GGPS gene have been found:
GGPS1 not been mapped due to severe segregation distortion
GGPS2 has been mapped to linkage group 2 of the B493xQAL map
Two copies of the PSY gene have been found:
PSY1 has been mapped to linkage group 3 of the B493xQAL map
PSY2 has been mapped to linkage group 8 of the B493xQAL map
One copy of the PDS gene has been found:
PDS has been mapped to linkage group 2 of the B493xQAL map
Two copies of the ZDS gene have been found:
ZDS1 has been mapped to linkage group 4 of the B493xQAL map
ZDS2 has been mapped to linkage group 5 of the B493xQAL map
One copy of the CRTISO gene has been found:
CRTISO has been mapped to linkage group 6 of the B493xQAL map
Two copies of the LCYB gene have been found:
LCYB1 has been mapped to linkage group 3 of the B493xQAL map
LCYB2 (CCS) has been mapped to linkage group 3 of the B493xQAL map
See also CCS
One copy of the LCYE gene has been found:
LCYE has been mapped to linkage group 9 of the B493xQAL map
Three copies of the CHXB gene have been found:
CHXB1 has been mapped to linkage group 3 of the B493xQAL map
CHXB2 has been mapped to linkage group 6 of the B493xQAL map
CHXB3 could not be mapped because no polymorphisms were found
One copy of the CHXE gene has been found:
CHXE has been mapped to linkage group 2 of the B493xQAL map
One copy of the ZEP gene has been found:
ZEP has been mapped to linkage group 5 of the B493xQAL map
One copy of the VDE gene has been found:
VDE has been mapped to linkage group 6 of the B493xQAL map
One copy of the CCS gene has been found:
CCS has been mapped to linkage group 3 of the B493xQAL map
See also LCYB2
Three copies of the NCED gene have been found:
NCED1 has been mapped to linkage group 2 of the B493xQAL map
NCED2 has been mapped to linkage group 2 of the B493xQAL map
NCED3 has been mapped to linkage group 6 of the B493xQAL map
Three copies of the CCD gene have been found:
CCD1 has been mapped to linkage group 3 of the B493xQAL map
CCD2 has been mapped to linkage group 1 of the B493xQAL map
CCD3 has been mapped to linkage group 3 of the B493xQAL map
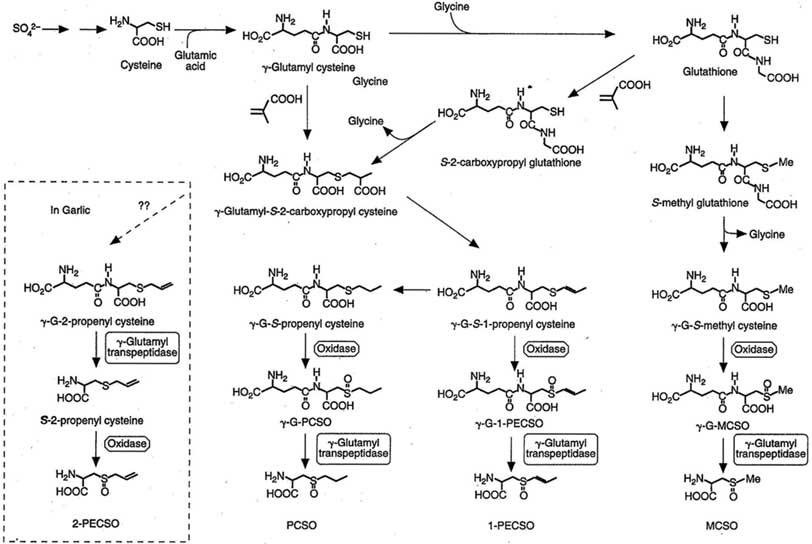
Flavor Precursors - This pathway is Figure 14.4 as published by Randle and Lancaster in Allium Crop Science: Recent Advances. Chapter 14. Sulphur Compounds in Alliums in Relation to Flavor Quality. CAB International, 2002 "Proposed biosynthesis of ACSOs and their intermediates"
No sequence information is available for any enzymes in this pathway, however, you may search for more recent information.
Click here to perform a search at NCBI for all sequences containing the term
Cysteine AND Oxidase limited to the Asparagales
Click here to perform a search at NCBI for all sequences containing the term
Glutamyl transpeptidase limited to the Asparagales
(These searches both returned no "No items found" in August 2009)
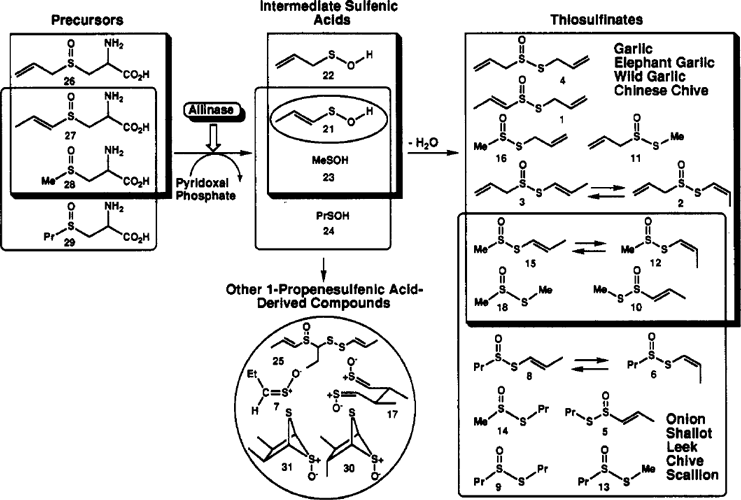
Alliinase - This pathway is Scheme II as published by Block et al. in J. Agric. Food Chem. Vol. 40 No. 12, 1992, pages 2418-2430.
This scheme depicts the different thiosulfinates and related sulfenic-acid derived compounds (7,17,25,30, and 31) found in extracts of nine Allium species. The upper, shadowed rectangles include allyl-group containing products from garlic, elephant garlic, wild garlic, and Chinese chive. The lower rectangles include propyl group-containing products from onion, shallot, leek, chive, and scallion. Thiosulfinates containing both methyl and l-propenyl groups are found in all Allium species examined and therefore are represented as lying within the confines of both sets of rectangles. This scheme also indicates the two types of precursors to the thiosulfinates, S-alk(en)yl cysteine S-oxides and sulfenic acids.
This is a clickable pathway, click on the enzyme or on the chemical to view more information
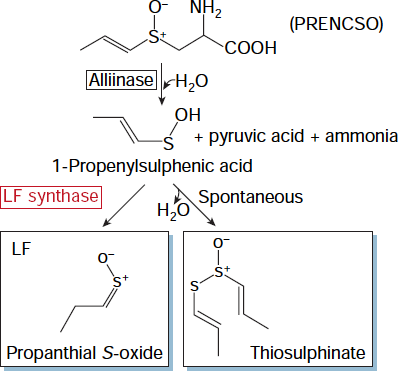
Lachrymatory Factor Synthase - This pathway was published by Imai et al. in Nature 419: 685. 2002. An onion enzyme that makes the eyes water
This is a clickable pathway, click on the enzyme or on the chemical to view more information
Key to chemical compounds shown in the pathways above
Catalyses the release of the S-alk(en)yl sulfoxide group from a S-alk(en)yl cysteine sulfoxide (ACSO)
Protein Data Bank structures
This page last modified Tuesday, 14-Dec-2010 12:32:56 CST