
This web site, https://www.vcru.wisc.edu/sdata, contains data from the laboratory of Philipp W. Simon, USDA-ARS Vegetable Crops Research Unit [Click here for our web page]
 |
USDA ARS VCRU Data Server This web site, https://www.vcru.wisc.edu/sdata, contains data from the laboratory of Philipp W. Simon, USDA-ARS Vegetable Crops Research Unit [Click here for our web page] |
This map was published in Theor Appl Genet (2000) 100:439–446
L.S. Boiteux, J.G. Belter, P.A. Roberts, P.W. Simon
RAPD linkage map of the genomic region encompassing the root-knot nematode (Meloidogyne javanica) resistance locus in carrot
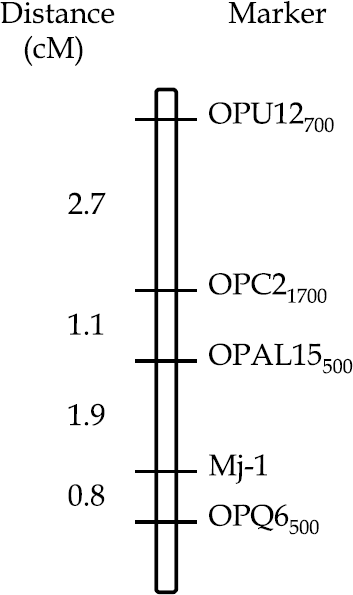
Abstract Inheritance studies have indicated that resistance to the root-knot nematode (Meloidogyne javanica) in carrot inbred line ‘Brasilia-1252’ is controlled by the action of one or two (duplicated) dominant gene(s) located at a single genomic region (designated the Mj-1 locus). A systematic search for randomly amplified polymorphic DNA (RAPD) markers linked to Mj-1 was carried out using bulked segregant analysis (BSA). Altogether 1000 ten-mer primers were screened with 69.1% displaying scorable amplicons. A total of approximately 2400 RAPD bands were examined. Four reproducible markers (OP-C21700, OP-Q6500, OP-U12700, and OP-AL15500) were identified, in coupling-phase linkage, flanking the Mj-1 region. The genetic distances between RAPD markers and the Mj-1 locus, estimated using an F2 progeny of 412 individuals from ‘Brasilia 1252’ × ‘B6274’, ranged from 0.8 to 5.7 cM . The two closest flanking markers (OP-Q6500 and OP-AL15500) encompassed a region of 2.7 cM . The frequency of these RAPD loci was evaluated in 121 accessions of a broadbased carrot germplasm collection. Only five entries (all resistant to M. javanica and genetically related to ‘Brasilia 1252’) exhibited the simultaneous presence of all four markers. An advanced line derived from the same cross, susceptible to M. javanica but relatively resistant to another root-knot nematode species (M. incognita), did not share three of the closest markers. These results suggest that at least some genes controlling resistance to M. incognita and M. javanica in ‘Brasilia 1252’ reside at distinct loci. The low number of markers suggests a reduced amount of genetic divergence between the parental lines at the region surrounding the target locus. Nevertheless, the low rate of recombination indicated these markers could be useful landmarks for positional cloning of the resistance gene(s). These RAPD markers could also be used to increase the Mj-1 frequency during recurrent selection cycles and in backcrossing programs to minimize ‘linkage drag’ in elite lines employed for the development of resistant F1 hybrids.
Details about individual markers are available by clicking on any of the markers on the map, or by going directly to the marker information page
The RAPD linkage map of the Mj-1 (M. javanica resistance) locus in carrot. The map population was composed by 412 F2 individuals from a cross between ‘Brasilia-1252’ (resistant parent) x ‘B6274’ (susceptible parent). The genetic distance in cM (Kosambi function) between pairs of markers is shown at the left and the RAPD marker designation is shown on the right.
 |
OPU12-700 |