
This web site, https://www.vcru.wisc.edu/sdata, contains data from the laboratory of Philipp W. Simon, USDA-ARS Vegetable Crops Research Unit [Click here for our web page]
 |
USDA ARS VCRU Data Server This web site, https://www.vcru.wisc.edu/sdata, contains data from the laboratory of Philipp W. Simon, USDA-ARS Vegetable Crops Research Unit [Click here for our web page] |
Merging Carrot Linkage Groups based on Conserved Dominant AFLP Markers in F2 Populations
Carlos A. F. Santos, Philipp W. Simon
J. Amer. Soc. Hort. Sci. 129(2):211-217. 2004
Abstract Markers were placed on linkage groups, ordered, and merged for two unrelated F2 populations of carrot (Daucus carota L.). Included were 277 and 242 dominant AFLP markers and 10 and eight co-dominant markers assigned to the nine linkage groups of Brasilia × HCM and B493 × QAL F2 populations, respectively. The merged linkage groups were based on two co-dominant markers and 28 conserved dominant AFLP markers (based upon sequence and size) shared by both populations. The average marker spacing was 4.8 to 5.5 cM in the four parental coupling phase maps. The average marker spacing in the six merged linkage groups was 3.75 cM with maximum gaps among linkage groups ranging from 8.0 to 19.8 cM. Gaps of a similar size were observed with the linkage coupling phase maps of the parents, indicating that linkage group integration did not double the bias which comes with repulsion phase mapping. Three out of nine linkage groups of carrot were not merged due to the absence of common markers. The six merged linkage groups incorporated similar numbers of AFLP fragments from the four parents, further indicating no significant increase in bias expected with repulsion phase linkage. While other studies have merged linkage maps with shared AFLPs of similar size, this is the first report to use shared AFLPs with highly conserved sequence to merge linkage maps in carrot. The genome coverage in this study is suitable to apply quantitative trait locus analysis and to construct a cross-validated consensus map of carrot, which is an important step toward an integrated map of carrot.
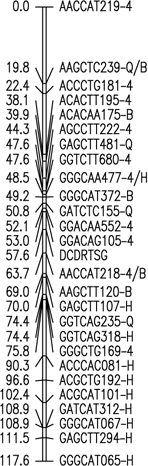
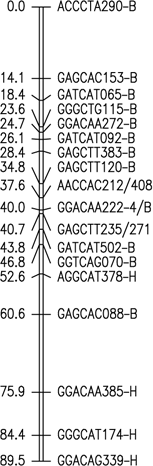
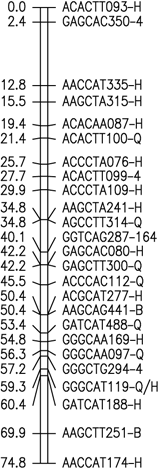
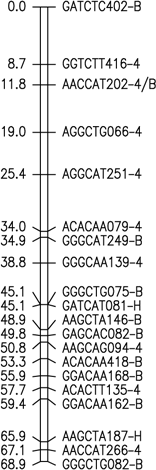
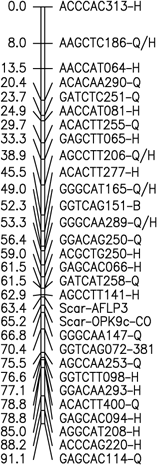
Fig. 1. Six merged linkage groups for Brasilia × HCM and B493 × QAL
F2 populations of carrot. AFLP fragments with / (e.g. AAGCTC239-Q/B) and two pair base sizes
(e.g. GAGCTT235/271) represent a conserved AFLP fragment between the two populations and a co-dominant
AFLP, respectively.
The map data for these maps (manually entered from the figures) is available as Mergedmap.txt
 |
 |
 |
 |
 |
 |
| 1 | 2 | 3 | 4 | 5 | 6 |
Table 1. Heterogeneity test (H) for recombination fraction (θ) per linkage group among common molecular markers used to merge linkage groups between the Brasilia (Bsb) × HCM and B493 × QAL F2 carrot populations (P).
| Pair | F2 | θ | LOD | Chi-square | Linkage group |
|
|---|---|---|---|---|---|---|
| P | H | |||||
| AACCAT218-4/B × AGGCTA108-4/H | Bsb × HCM | 0.3033 | 1.42 | 3.70 | LG1 | |
| B493 × QAL | 0.1340 | 14.47 | 1.69 | 5.39* | LG1 | |
| AACCAT218-4/B × GAGCAC444-4/H | Bsb × HCM | 0.2005 | 9.02 | 0.07 | LG1 | |
| B493 × QAL | 0.2401 | 2.36 | 0.19 | 0.26 NS | LG1 | |
| AACCAT218-4/B × DCDRTSG | Bsb × HCM | 0.0658 | 23.19 | 1.26 | LG1 | |
| B493 × QAL | 0.1230 | 16.49 | 1.05 | 2.30NS | LG1 | |
| AACCAT218-4/B × GATCTC181-4/H | Bsb × HCM | 0.1937 | 2.47 | 0.17 | LG1 | |
| B493 × QAL | 0.1561 | 12.14 | 0.04 | 0.22NS | LG1 | |
| AGGCTA108-4/H × DCDRTSG | Bsb × HCM | 0.3194 | 2.01 | 7.43 | LG1 | |
| B493 × QAL | 0.1044 | 18.73 | 5.70 | 13.12** | LG1 | |
| AGGCTA108-4/H × GAGCAC444-4/H | Bsb × HCM | 0.0928 | 19.58 | 1.21 | LG1 | |
| B493 × QAL | 0.1704 | 8.57 | 1.43 | 2.64 NS | LG1 | |
| AGGCTA108-4/H × GATCTC181-4/H | Bsb × HCM | 0.0722 | 16.43 | 2.24 | LG1 | |
| B493 × QAL | 0.0133 | 32.85 | 2.67 | 4.91* | LG1 | |
| GAGCAC444-4/H × GATCTC181-4/H | Bsb × HCM | 0.0658 | 23.15 | 0.87 | LG1 | |
| B493 × QAL | 0.1229 | 11.44 | 1.04 | 1.91 NS | LG1 | |
| GAGCAC444-4/H × DCDRTSG | Bsb × HCM | 0.1870 | 9.79 | 1.95 | LG1 | |
| B493 × QAL | 0.3247 | 2.01 | 2.49 | 4.44* | LG1 | |
| DCDRTSG × GATCTC181-4/H | Bsb × HCM | 0.2902 | 2.77 | 5.19 | LG1 | |
| B493 × QAL | 0.1102 | 17.07 | 4.11 | 9.30** | LG1 | |
| GGACAA222-4/B × GGGCAT282-4/H | Bsb × HCM | 0.2551 | 1.95 | 4.45 | LG2 | |
| B493 × QAL | 0.0805 | 18.82 | 2.14 | 6.59* | LG2 | |
| GGACAA222-4/B × AAGCAG075-4/H | Bsb × HCM | 0.3509 | 0.85 | 0.80 | LG2 | |
| B493 × QAL | 0.2561 | 4.58 | 0.43 | 1.23 NS | LG2 | |
| GGGCAT282-4/H × GGACAG272-4/H | Bsb × HCM | 0.3339 | 1.05 | 0.08 | LG2 | |
| B493 × QAL | 0.3676 | 1.09 | 0.05 | 0.13 NS | LG2 | |
| GGGCAT282-4/H × AAGCAG075-4/H | Bsb × HCM | 0.2824 | 3.23 | 0.15 | LG2 | |
| B493 × QAL | 0.2435 | 4.85 | 0.15 | 0.31 NS | LG2 | |
| AAGCAG075-4/H × GGACAG272-4/H | Bsb × HCM | 0.2334 | 4.64 | 1.51 | LG2 | |
| B493 × QAL | 0.1304 | 12.07 | 1.39 | 2.91 NS | LG2 | |
| GGACAG259-4/B × ACACAA142-4/H | Bsb × HCM | 0.2477 | 1.68 | 0.39 | LG3 | |
| B493 × QAL | 0.1834 | 8.08 | 0.15 | 0.54 NS | LG3 | |
| GGACAG259-4/B × AGCCAA136-Q/H | Bsb × HCM | 0.3835 | 0.53 | 0.84 | LG3 | |
| B493 × QAL | 0.2584 | 2.56 | 0.83 | 1.67 NS | LG3 | |
| ACACAA142-4/H × AGCCAA136-Q/H | Bsb × HCM | 0.3984 | 0.67 | 0.98 | LG3 | |
| B493 × QAL | 0.2564 | 2.54 | 1.66 | 2.64 NS | LG3 | |
| AACCAT100-4/H × AAGCAG097-4/B | Bsb × HCM | 0.2256 | 1.97 | 0.86 | LG4 | |
| B493 × QAL | 0.1434 | 15.03 | 0.22 | 1.09 NS | LG4 | |
| AACCAT100-4/H × GGGCAT119-Q/H | Bsb × HCM | 0.3308 | 1.83 | 0.01 | LG4 | |
| B493 × QAL | 0.3224 | 2.13 | 0.01 | 0.01 NS | LG4 | |
| AAGCAG097-4/B × GGGCAT119-Q/H | Bsb × HCM | 0.3200 | 1.20 | 0.27 | LG4 | |
| B493 × QAL | 0.2527 | 2.80 | 0.24 | 0.51 NS | LG4 | |
| AACCAT202-4/B × ACACAA106-Q/B | Bsb × HCM | 0.0979 | 15.43 | 0.23 | LG5 | |
| B493 × QAL | 0.0000 | 4.82 | 2.80 | 3.04 NS | LG5 | |
| AACCAT202-4/B × ACACTT153-4/H | Bsb × HCM | 0.2567 | 2.01 | 0.01 | LG5 | |
| B493 × QAL | 0.2465 | 5.22 | 0.00 | 0.01 NS | LG5 | |
| AACCAT202-4/B × GGGCAA229-4/H | Bsb × HCM | 0.4136 | 0.34 | 0.01 | LG5 | |
| B493 × QAL | 0.4228 | 0.35 | 0.01 | 0.01 NS | LG5 | |
| ACACAA106-Q/B × ACACTT153-4/H | Bsb × HCM | 0.2442 | 2.34 | 0.02 | LG5 | |
| B493 × QAL | 0.2206 | 2.33 | 0.03 | 0.05 NS | LG5 | |
| ACACAA106-Q/B × GGGCAA229-4/H | Bsb × HCM | 0.3395 | 0.99 | 0.74 | LG5 | |
| B493 × QAL | 0.4418 | 0.21 | 0.44 | 1.18 NS | LG5 | |
| ACACTT153-4/H × GGGCAA229-4/H | Bsb × HCM | 0.2869 | 3.56 | 0.81 | LG5 | |
| B493 × QAL | 0.2033 | 7.23 | 0.86 | 1.66 NS | LG5 | |
| AGCCTT206-Q/H × GATCTC386-4/B | Bsb × HCM | 0.3503 | 0.70 | 1.10 | LG6 | |
| B493 × QAL | 0.1834 | 3.10 | 1.35 | 2.45 NS | LG6 | |
| AGCCTT206-Q/H × Scar-AFLP3 | Bsb × HCM | 0.2191 | 5.37 | 0.14 | LG6 | |
| B493 × QAL | 0.1915 | 11.62 | 0.08 | 0.22 NS | LG6 | |
| AGCCTT206-Q/H × ACACAA319-4/H | Bsb × HCM | 0.1808 | 8.08 | 0.59 | LG6 | |
| B493 × QAL | 0.0000 | 5.21 | 5.62 | 6.21* | LG6 | |
| AGCCTT206-Q/H × GGGCAT165-Q/H | Bsb × HCM | 0.1420 | 10.27 | 0.20 | LG6 | |
| B493 × QAL | 0.1157 | 20.08 | 0.13 | 0.32 NS | LG6 | |
| AGCCTT206-Q/H × GGTCTT350-4/H | Bsb × HCM | 0.1524 | 9.16 | 0.40 | LG6 | |
| B493 × QAL | 0.0000 | 4.52 | 4.11 | 4.51* | LG6 | |
| AGCCTT206-Q/H × GGGCAA289-Q/H | Bsb × HCM | 0.1637 | 7.87 | 0.35 | LG6 | |
| B493 × QAL | 0.1260 | 18.61 | 0.2 | 0.55 NS | LG6 | |
| GATCTC386-4/B × Scar-AFLP3 | Bsb × HCM | 0.0552 | 24.71 | 0.01 | LG6 | |
| B493 × QAL | 0.0605 | 22.45 | 0.01 | 0.03 NS | LG6 | |
| GATCTC386-4/B × ACACAA319-4/H | Bsb × HCM | 0.1539 | 4.21 | 0.90 | LG6 | |
| B493 × QAL | 0.2445 | 4.69 | 0.38 | 1.27 NS | LG6 | |
| GATCTC386-4/B × GGGCAT165-Q/H | Bsb × HCM | 0.2374 | 2.35 | 0.42 | LG6 | |
| B493 × QAL | 0.1519 | 4.39 | 0.46 | 0.88 NS | LG6 | |
| GATCTC386-4/B × GGTCTT350-4/H | Bsb × HCM | 0.3220 | 1.02 | 3.51 | LG6 | |
| B493 × QAL | 0.1389 | 11.45 | 1.75 | 5.25* | LG6 | |
| GATCTC386-4/B × GGGCAA289-Q/H | Bsb × HCM | 0.2729 | 1.45 | 0.69 | LG6 | |
| B493 × QAL | 0.1553 | 4.04 | 0.69 | 1.38 NS | LG6 | |
| Scar-AFLP3 × ACACAA319-4/H | Bsb × HCM | 0.1736 | 10.66 | 0.10 | LG6 | |
| B493 × QAL | 0.1990 | 8.03 | 0.11 | 0.21 NS | LG6 | |
| Scar-AFLP3 × GGGCAT165-Q/H | Bsb × HCM | 0.1531 | 10.35 | 0.50 | LG6 | |
| B493 × QAL | 0.1108 | 20.52 | 0.36 | 0.86 NS | LG6 | |
| Scar-AFLP3 × GGTCTT350-4/H | Bsb × HCM | 0.1608 | 9.03 | 1.48 | LG6 | |
| B493 × QAL | 0.0808 | 19.12 | 1.40 | 2.88 NS | LG6 | |
| Scar-AFLP3 × GGGCAA289-Q/H | Bsb × HCM | 0.1144 | 12.55 | 0.16 | LG6 | |
| B493 × QAL | 0.0927 | 23.33 | 0.10 | 0.27 NS | LG6 | |
| ACACAA319-4/H × GGGCAT165-Q/H | Bsb × HCM | 0.0667 | 20.16 | 4.03 | LG6 | |
| B493 × QAL | 0.2557 | 2.64 | 5.85 | 9.88** | LG6 | |
| ACACAA319-4/H × GGTCTT350-4/H | Bsb × HCM | 0.1573 | 10.17 | 0.03 | LG6 | |
| B493 × QAL | 0.1439 | 11.52 | 0.03 | 0.06 NS | LG6 | |
| ACACAA319-4/H × GGGCAA289-Q/H | Bsb × HCM | 0.1252 | 13.08 | 0.58 | LG6 | |
| B493 × QAL | 0.2139 | 3.21 | 1.12 | 1.70 NS | LG6 | |
| GGGCAT165-Q/H × GGTCTT350-4/H | Bsb × HCM | 0.0790 | 16.74 | 0.27 | LG6 | |
| B493 × QAL | 0.0000 | 5.92 | 2.65 | 2.95 NS | LG6 | |
| GGGCAT165-Q/H × GGGCAA289-Q/H | Bsb × HCM | 0.0713 | 17.34 | 0.14 | LG6 | |
| B493 × QAL | 0.0555 | 29.74 | 0.09 | 0.24 NS | LG6 | |
| GGTCTT350-4/H × GGGCAA289-Q/H | Bsb × HCM | 0.0713 | 17.34 | 0.14 | LG6 | |
| B493 × QAL | 0.055 | 29.74 | 0.09 | 0.24 NS | LG6 | |
**, * and NS significant at the p-values of 1%, 5% and non-significant, respectively, by chi-square test
Supplementary Table A. List of the two co-dominant markers and 28 conserved dominant AFLP markers used for merging the maps, and the corresponding map location of these markers in the previously published B493×QAL and BSB×HCM maps
| Marker Name | Merged Group | B493×QAL Location | BSB×HCM Location | |
|---|---|---|---|---|
| DCDRTSG | 1@57.6 | LG1 | LG1 | |
| Scar-AFLP3 | 6@63.4 | LG6 | LG6 | |
| AACCAT100-4/H | not in merged map | LG4 | LG4 | |
| AACCAT202-4/B | 5@11.8 | LG5 | LG5 | |
| AACCAT218-4/B | 1@63.7 | LG1 | LG1 | |
| AACCAT315-Q/H | not in merged map | not mapped | LG8 | |
| AAGCAG075-4/H | not in merged map | LG2 | LG2 | |
| AAGCAG097-4/B | not in merged map | LG4 | not mapped | |
| AAGCAG134-Q/B | not in merged map | LG7 | LG7 | |
| AAGCTC186-Q/H | 3@65.1 &6@ 8.0 | LG3 ≠ | LG6 | |
| AAGCTC239-Q/B | 1@19.8 | LG1 | LG8 | |
| ACACAA106-Q/B | not in merged map | LG5 | LG5 | |
| ACACAA142-4/H | not in merged map | not mapped | LG3 | |
| ACACAA319-4/H | not in merged map | LG6 | LG6 | |
| ACACTT153-4/H | not in merged map | LG5 | LG5 | |
| AGCCAA136-Q/H | 3@21.0 | LG3 | LG3 | |
| AGCCTT206-Q/H | 6@38.9 | LG6 | LG6 | |
| AGGCTA108-4/H | not in merged map | LG1 | LG1 | |
| GAGCTT203-4/B | not in merged map | not mapped | LG8 | |
| GATCTC181-4/H | not in merged map | LG1 | LG1 | |
| GATCTC386-4/B | not in merged map | LG6 | LG6 | |
| GGACAA222-4/B | 2@40.0 | LG2 | LG2 | |
| GGACAG259-4/B | not in merged map | LG3 | LG3 | |
| GGACAG272-4/H | not in merged map | not mapped | not mapped | |
| GGGCAA229-4/H | not in merged map | LG5 | LG5 | |
| GGGCAA289-Q/H | 6@53.3 | LG6 | LG6 | |
| GGGCAA477-4/H | 1@48.5 | LG1 ≠ | LG7 | |
| GGGCAT119-Q/H | 4@59.3 | LG4 | LG4 | |
| GGGCAT165-Q/H | 6@49.0 | LG6 | LG6 | |
| GGGCAT282-4/H | not in merged map | LG2 | LG2 | |
| GGTCTT350-4/H | not in merged map | LG6 | LG6 | |
| Others not used | ||||
| GGACAA272-Q/B | LG2 | not scored | ||
| (shown as GGACAA272-Q in map) | ||||
| GAGCAC444-4/H | LG1 | LG1 | ||